ARTADE2 予測遺伝子モデル データベースTAIR_gene_model (on ARTADE2DB) (ver. TAIR9)Genes with tiling-array support (Annotated)
AT1G29090.1http://metadb.riken.jp/db/SciNetS_ria227i/cria227s2ria227u122909001000i
Other supporting information
| Gene Model | |
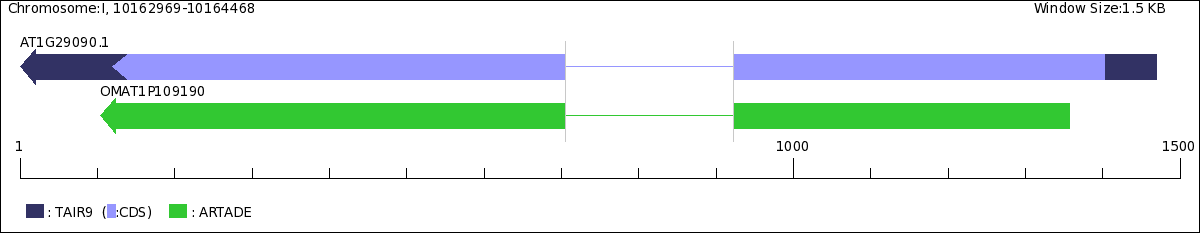 | |
| Correlation Plot | |
 | |
Expression profile (Values are plotted in Log(2) values.)
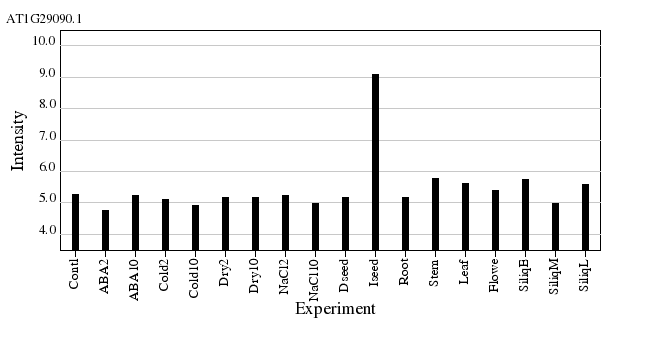
Genes with related expresssion profiles.
| Positively Correlated Genes | ||||||
|---|---|---|---|---|---|---|
| Gens | PCC | Definition | Overlap gene | Definition | Overlap gene(antisense) | Definition |
| AT1G29080.1 | 0.99819 | peptidase C1A papain family protein | OMAT1P109180 | - | - | - |
| AT1G60090.1 | 0.997978 | BGLU4 (BETA GLUCOSIDASE 4) | OMAT1P016400 | - | - | - |
| AT1G43780.1 | 0.997773 | scpl44 (serine carboxypeptidase-like 44) | OMAT1P012520 | - | - | - |
| AT5G05290.1 | 0.997632 | ATEXPA2 (ARABIDOPSIS THALIANA EXPANSIN A2) | OMAT5P001690 | - | OMAT5P101345 | - |
| AT4G36880.1 | 0.997506 | CP1 (CYSTEINE PROTEINASE1) | OMAT4P111050 | - | OMAT4P012690 | - |
| AtsnoR67 | 0.995689 | AtsnoR67 | - | - | - | - |
| AtsnoR53Y | 0.995469 | AtsnoR53Y | OMAT1P114020 | - | - | - |
| AT2G26040.1 | 0.994587 | Bet v I allergen family protein | OMAT2P104870 | - | - | - |
| AT3G04800.1 | 0.99445 | ATTIM23-3 | OMAT3P101550 | - | - | - |
| AT5G24990.1 | 0.994105 | unknown protein | - | - | - | - |
| Negatively Correlated Genes | ||||||
| Gens | PCC | Definition | Overlap gene | Definition | Overlap gene(antisense) | Definition |
| AT1G51402.1 | -0.695445 | unknown protein | OMAT1P014180 | - | OMAT1P112450 | - |
| AT4G17170.1 | -0.651939 | RABB1C (ARABIDOPSIS RAB GTPASE HOMOLOG B1C) | OMAT4P104230 | - | - | - |
| AT3G01202.1 | -0.647183 | other RNA | - | - | OMAT3P100100 | - |
| AT5G44100.1 | -0.642983 | ckl7 (Casein Kinase I-like 7) | - | - | - | - |
| AT4G20260.4 | -0.626609 | DREPP plasma membrane polypeptide family protein | OMAT4P006170 | - | - | - |
| AT1G20620.1 | -0.626204 | CAT3 (CATALASE 3) | OMAT1P007530,OMAT1P007540 | [OMAT1P007530]-, [OMAT1P007540]- | OMAT1P106670 | - |
| AT2G05600.1 | -0.598701 | FUNCTIONS IN: molecular_function unknown | - | - | - | - |
| AT5G12980.1 | -0.587684 | rcd1-like cell differentiation protein, putative | - | - | - | - |
| AT3G49460.1 | -0.585708 | 60S acidic ribosomal protein-related | - | - | - | - |
| AT1G37007.1 | -0.57739 | unknown protein | - | - | - | - |
Get whole results
Over-Representation Analysis Result
| p-value | <= 2.42e-12 | :20 terms with high significance | |
| 2.42e-12 < | p-value | <= 8.76e-06 | :With considering multiple testing correction; p <= 1.00e-02 / 1142 |
| 8.76e-06 < | p-value | <= 1.00e-02 |
| Type of term (*1) | Depth of the term in ontology tree | ID/Term | Description | Number of genes | Over-Representative rate (*2) | p-value (*3) | PosMed p-value (*4) (Link to PosMed) | Found on gene annotation |
|---|---|---|---|---|---|---|---|---|
| B | 4 | GO:0042254 | ribosome biogenesis | 26/200 | 20.30 | 8.29e-28 | - | no |
| B | 3 | GO:0022613 | ribonucleoprotein complex biogenesis | 26/200 | 19.65 | 2.03e-27 | - | no |
| B | 5 | GO:0006364 | rRNA processing | 16/200 | 25.58 | 7.67e-20 | - | no |
| B | 5 | GO:0009451 | RNA modification | 14/200 | 18.48 | 1.64e-15 | - | no |
| B | 5 | GO:0006396 | RNA processing | 20/200 | 9.04 | 1.09e-14 | - | no |
| B | 4 | GO:0010467 | gene expression | 52/200 | 2.55 | 4.63e-11 | 1.29E-17 | no |
| B | 5 | GO:0006412 | translation | 28/200 | 3.86 | 2.27e-10 | - | no |
| B | 3 | GO:0043170 | macromolecule metabolic process | 73/200 | 1.89 | 4.14e-09 | - | yes |
| B | 4 | GO:0044260 | cellular macromolecule metabolic process | 64/200 | 1.82 | 2.41e-07 | - | no |
| B | 3 | GO:0044238 | primary metabolic process | 80/200 | 1.61 | 6.84e-07 | - | yes |
| B | 5 | GO:0016070 | RNA metabolic process | 24/200 | 2.82 | 1.50e-06 | - | no |
| B | 4 | GO:0019538 | protein metabolic process | 45/200 | 1.95 | 3.39e-06 | - | yes |
| B | 3 | GO:0044237 | cellular metabolic process | 70/200 | 1.48 | 9.86e-05 | - | no |
| B | 5 | GO:0044267 | cellular protein metabolic process | 36/200 | 1.77 | 2.60e-04 | - | no |
| B | 5 | GO:0034645 | cellular macromolecule biosynthetic process | 37/200 | 1.72 | 3.81e-04 | - | no |
| B | 4 | GO:0009059 | macromolecule biosynthetic process | 37/200 | 1.71 | 4.04e-04 | - | no |
| B | 5 | GO:0090304 | nucleic acid metabolic process | 28/200 | 1.86 | 4.99e-04 | - | no |
| B | 4 | GO:0006139 | nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 30/200 | 1.78 | 7.17e-04 | - | no |
| B | 5 | GO:0006508 | proteolysis | 11/200 | 2.62 | 1.12e-03 | - | yes |
| B | 4 | GO:0034641 | cellular nitrogen compound metabolic process | 30/200 | 1.54 | 6.35e-03 | - | no |
| B | 4 | GO:0044249 | cellular biosynthetic process | 40/200 | 1.42 | 7.89e-03 | - | no |
| B | 3 | GO:0006807 | nitrogen compound metabolic process | 30/200 | 1.51 | 8.35e-03 | - | no |
| C | 3 | GO:0030529 | ribonucleoprotein complex | 40/200 | 11.47 | 1.22e-31 | - | no |
| C | 3 | GO:0043228 | non-membrane-bounded organelle | 38/200 | 6.38 | 7.92e-21 | - | no |
| C | 4 | GO:0043232 | intracellular non-membrane-bounded organelle | 38/200 | 6.38 | 7.92e-21 | - | no |
| C | 4 | GO:0005840 | ribosome | 26/200 | 10.15 | 9.11e-20 | - | no |
| C | 4 | GO:0033279 | ribosomal subunit | 20/200 | 12.32 | 2.18e-17 | - | no |
| C | 5 | GO:0015934 | large ribosomal subunit | 14/200 | 15.21 | 3.07e-14 | - | no |
| C | 3 | GO:0044424 | intracellular part | 96/200 | 1.88 | 1.92e-12 | - | no |
| C | 5 | GO:0005730 | nucleolus | 16/200 | 9.33 | 2.02e-12 | - | no |
| C | 3 | GO:0005622 | intracellular | 98/200 | 1.84 | 3.88e-12 | - | no |
| C | 5 | GO:0022626 | cytosolic ribosome | 16/200 | 8.69 | 6.28e-12 | - | no |
| C | 3 | GO:0044446 | intracellular organelle part | 43/200 | 3.00 | 1.77e-11 | - | no |
| C | 3 | GO:0044422 | organelle part | 43/200 | 3.00 | 1.82e-11 | - | no |
| C | 4 | GO:0070013 | intracellular organelle lumen | 18/200 | 6.64 | 4.21e-11 | - | no |
| C | 3 | GO:0043233 | organelle lumen | 18/200 | 6.62 | 4.37e-11 | - | no |
| C | 5 | GO:0044445 | cytosolic part | 13/200 | 8.68 | 4.14e-10 | - | no |
| C | 5 | GO:0031981 | nuclear lumen | 16/200 | 6.62 | 4.39e-10 | - | no |
| C | 3 | GO:0043229 | intracellular organelle | 81/200 | 1.75 | 1.22e-08 | - | no |
| C | 3 | GO:0044464 | cell part | 130/200 | 1.42 | 1.34e-08 | - | yes |
| C | 4 | GO:0044428 | nuclear part | 16/200 | 4.81 | 4.95e-08 | - | no |
| C | 4 | GO:0044444 | cytoplasmic part | 63/200 | 1.81 | 3.22e-07 | - | no |
| C | 4 | GO:0005737 | cytoplasm | 65/200 | 1.74 | 9.99e-07 | - | no |
| C | 5 | GO:0005739 | mitochondrion | 19/200 | 2.97 | 7.44e-06 | - | no |
| C | 5 | GO:0005829 | cytosol | 14/200 | 3.38 | 2.02e-05 | - | no |
| C | 4 | GO:0043231 | intracellular membrane-bounded organelle | 66/200 | 1.50 | 1.12e-04 | - | no |
| C | 3 | GO:0043227 | membrane-bounded organelle | 66/200 | 1.50 | 1.12e-04 | - | no |
| C | 5 | GO:0005634 | nucleus | 26/200 | 1.66 | 3.84e-03 | - | no |
| M | 3 | GO:0003735 | structural constituent of ribosome | 26/200 | 12.01 | 1.15e-21 | - | no |
| M | 3 | GO:0016787 | hydrolase activity | 26/200 | 1.66 | 3.78e-03 | - | no |
| PS | 4 | PO:0000037 | shoot apex | 118/200 | 1.37 | 1.92e-06 | - | no |
| PS | 3 | PO:0009005 | root | 121/200 | 1.34 | 3.70e-06 | - | no |
| PS | 5 | PO:0009052 | pedicel | 106/200 | 1.30 | 1.58e-04 | - | no |
| PS | 4 | PO:0009009 | embryo | 119/200 | 1.23 | 5.74e-04 | - | no |
| PS | 5 | PO:0020038 | petiole | 98/200 | 1.29 | 6.24e-04 | - | no |
| PS | 3 | PO:0009010 | seed | 120/200 | 1.23 | 6.34e-04 | - | no |
| PS | 4 | PO:0009001 | fruit | 120/200 | 1.22 | 8.14e-04 | - | no |
| PS | 3 | PO:0006342 | infructescence | 120/200 | 1.22 | 8.14e-04 | - | no |
| PS | 3 | PO:0009031 | sepal | 113/200 | 1.22 | 1.35e-03 | - | no |
| PS | 4 | PO:0009025 | leaf | 112/200 | 1.22 | 1.76e-03 | - | no |
| PS | 4 | PO:0009049 | inflorescence | 124/200 | 1.18 | 2.31e-03 | - | no |
| PS | 3 | PO:0009032 | petal | 107/200 | 1.22 | 2.36e-03 | - | no |
| PS | 5 | PO:0009046 | flower | 123/200 | 1.18 | 2.75e-03 | - | no |
| PS | 4 | PO:0009026 | sporophyll | 109/200 | 1.21 | 2.80e-03 | - | no |
| PS | 3 | PO:0009006 | shoot | 127/200 | 1.17 | 3.32e-03 | - | no |
| PS | 3 | PO:0006001 | phyllome | 121/200 | 1.18 | 3.48e-03 | - | no |
| PS | 5 | PO:0009028 | microsporophyll | 102/200 | 1.22 | 3.88e-03 | - | no |
| PS | 5 | PO:0009027 | megasporophyll | 102/200 | 1.21 | 4.05e-03 | - | no |
| PS | 5 | PO:0008037 | seedling | 106/200 | 1.20 | 4.75e-03 | - | no |
| PS | 5 | PO:0008034 | leaf whorl | 112/200 | 1.19 | 4.89e-03 | - | no |
| PS | 4 | PO:0008033 | phyllome whorl | 112/200 | 1.19 | 4.89e-03 | - | no |
| PG | 5 | PO:0001081 | F mature embryo stage | 111/200 | 1.40 | 2.43e-06 | - | no |
| PG | 5 | PO:0001078 | E expanded cotyledon stage | 113/200 | 1.36 | 6.82e-06 | - | no |
| PG | 5 | PO:0004507 | D bilateral stage | 109/200 | 1.32 | 5.38e-05 | - | no |
| PG | 4 | PO:0007631 | embryo development stages | 114/200 | 1.27 | 2.57e-04 | - | no |
| PG | 3 | PO:0001170 | seed development stages | 114/200 | 1.26 | 3.16e-04 | - | no |
| PG | 3 | PO:0007134 | A vegetative growth | 106/200 | 1.25 | 9.48e-04 | - | no |
| PG | 5 | PO:0007604 | corolla developmental stages | 118/200 | 1.20 | 2.47e-03 | - | no |
| PG | 5 | PO:0001185 | C globular stage | 101/200 | 1.23 | 2.70e-03 | - | no |
| PG | 5 | PO:0007133 | leaf production | 102/200 | 1.21 | 4.34e-03 | - | no |
| PG | 4 | PO:0007112 | 1 main shoot growth | 102/200 | 1.21 | 4.38e-03 | - | no |
| PG | 4 | PO:0007600 | 3 floral organ development stages | 118/200 | 1.16 | 9.07e-03 | - | no |
| PG | 4 | PO:0007616 | 4 anthesis | 113/200 | 1.17 | 9.49e-03 | - | no |
| KW | 0 | ribosomal | - | 31/200 | 10.27 | 2.27e-23 | - | no |
| KW | 0 | ribosome | - | 28/200 | 10.61 | 1.14e-21 | - | no |
| KW | 0 | constituent | - | 26/200 | 9.28 | 9.03e-19 | - | no |
| KW | 0 | structural | - | 26/200 | 7.42 | 2.46e-16 | - | no |
| KW | 0 | translation | - | 29/200 | 5.85 | 3.02e-15 | - | no |
| KW | 0 | subunit | - | 31/200 | 4.23 | 2.42e-12 | - | no |
| KW | 0 | large | - | 14/200 | 9.13 | 5.11e-11 | - | no |
| KW | 0 | cytosolic | - | 16/200 | 7.47 | 6.76e-11 | - | no |
| KW | 0 | nucleolus | - | 13/200 | 9.56 | 1.16e-10 | - | no |
| KW | 0 | mitochondrial | - | 14/200 | 6.21 | 1.02e-08 | - | no |
| KW | 0 | biogenesis | - | 10/200 | 7.36 | 1.41e-07 | - | no |
| KW | 0 | intracellular | - | 15/200 | 2.91 | 6.91e-05 | - | no |
| KW | 0 | peptidase | - | 11/200 | 2.76 | 7.21e-04 | 0 | yes |
| KW | 0 | terminal | - | 31/200 | 1.61 | 3.00e-03 | - | yes |
| KW | 0 | conserved | - | 23/200 | 1.70 | 4.63e-03 | - | no |
| KW | 0 | nucleotide | - | 13/200 | 2.04 | 5.02e-03 | - | no |
| KW | 0 | mitochondrion | - | 11/200 | 2.01 | 9.51e-03 | - | no |
| (*1) | [B]:Biological process(Gene ontology), [C]:Cellular component(Gene ontology), [M]:Molecular function(Gene ontology), [PS]:Plant Structure(Plant ontology), [PG]:Growth and developmental stages(Plant ontology), [KW]:words found in gene description. |
| (*2) | ([# of genes with the term] / [# of sampling (200)]) / ([# of genes with the term among whole genes] / [# of whole genes]) |
| (*3) | P-values were calculated on hypergeometric distribution in which we found n genes with a annotation term during 200 highly correlated genes, while we had N genes with the term in the whole genes. |
| (*4) | PosMed is a system which serve a p-values showing a relationship between the gene and the annotation term based on literature information and Gene-Gene interaction suchas co-expression or protein-protein interactions. |
Top Page


